Drop us a line
If you are interested in the development of a custom solution — send us the message and we'll schedule a talk about it.
A Biotechnology Corporation is engaged in providing access to genomic sequence information and conducting biological research. In order to beat off the challenge associated with short sequence searching, the company turned to Azati with the request to optimize the BLAST algorithm they use.
Today, with the advent of biotechnology, we can safely say that modern medicine is literally on the verge of a real revolution, which consists in the fact that instead of mass therapy, the patient will be able to receive personalized therapy modeled on a specific clinical situation.
Biotechnology, in its current formative stage, focuses on diseases for which there are currently no adequate effective therapies. In this case, we are talking about diseases that appear with age. Among them: malignant neoplasms, arthritis and neurodegenerative diseases (Alzheimer’s and Parkinson’s diseases). In addition, biotechnology offers effective therapeutic solutions in the treatment of hepatitis, multiple sclerosis and orphan diseases.
Our customer is a biotech company that turned to Azati to improve their processes.
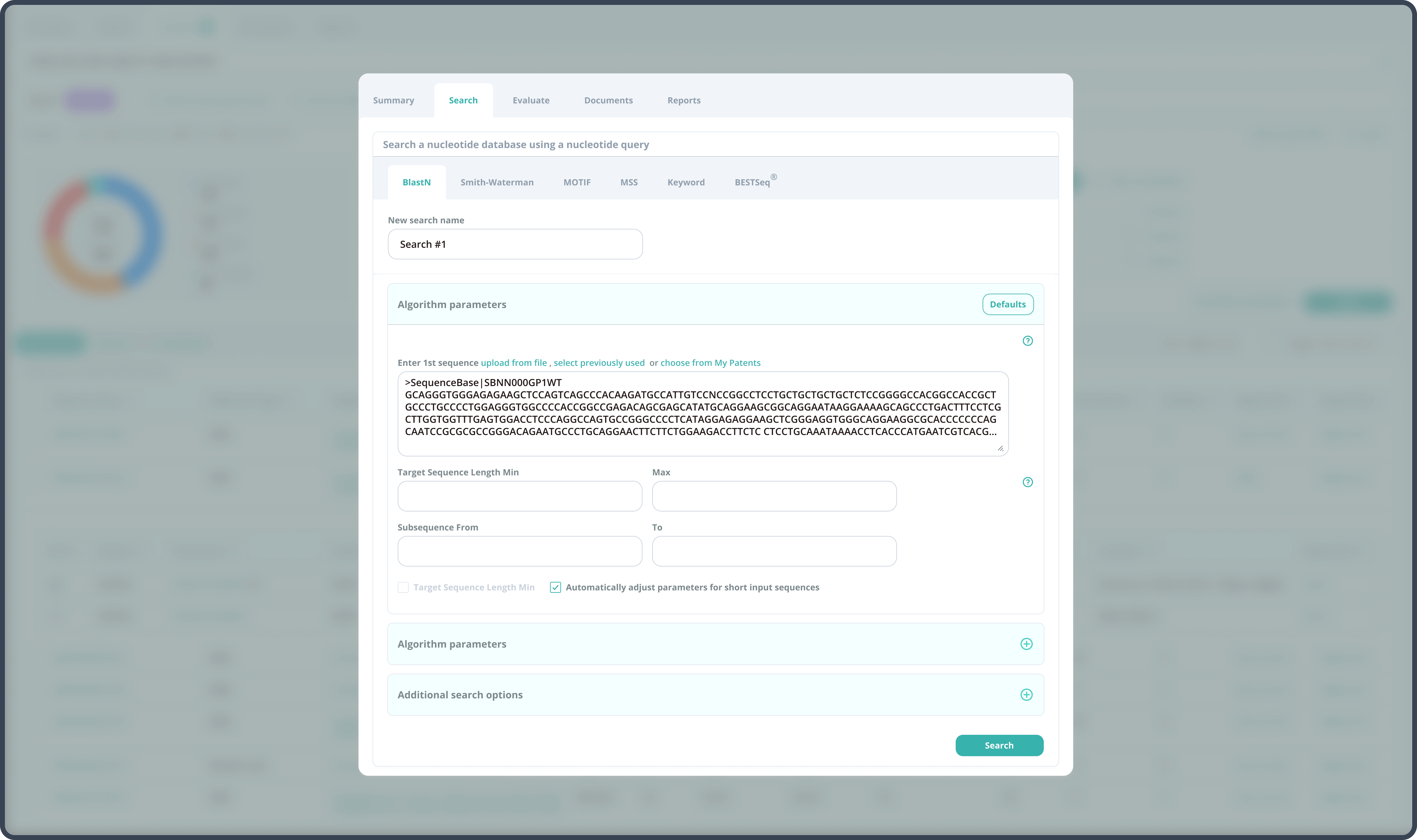
One of the research teams of our client company is dedicated to working with primers, which are short strands of DNA/ RNA used in the polymerase chain reaction or hybridization. For biological sequence comparison the researchers use BLAST algorithm.
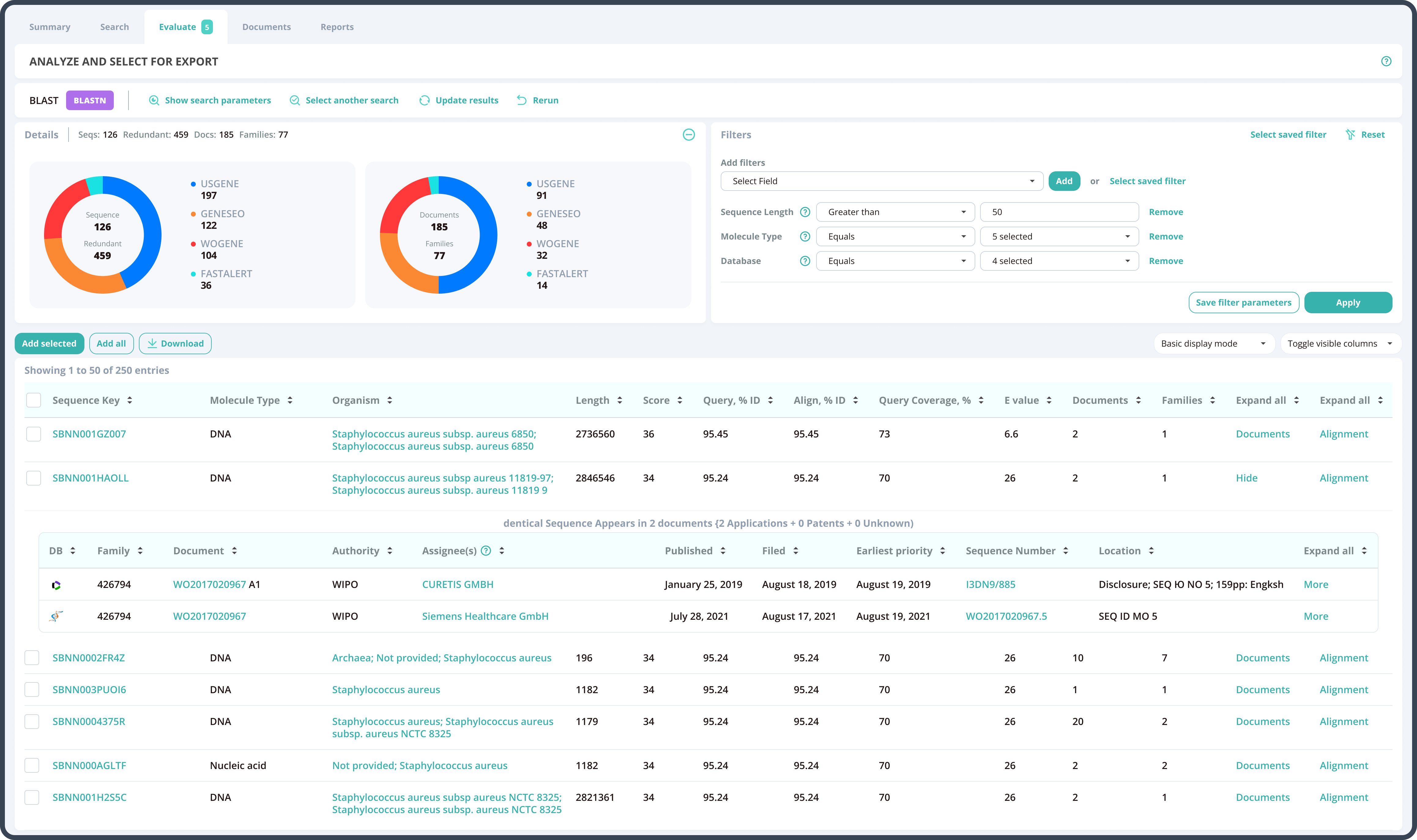
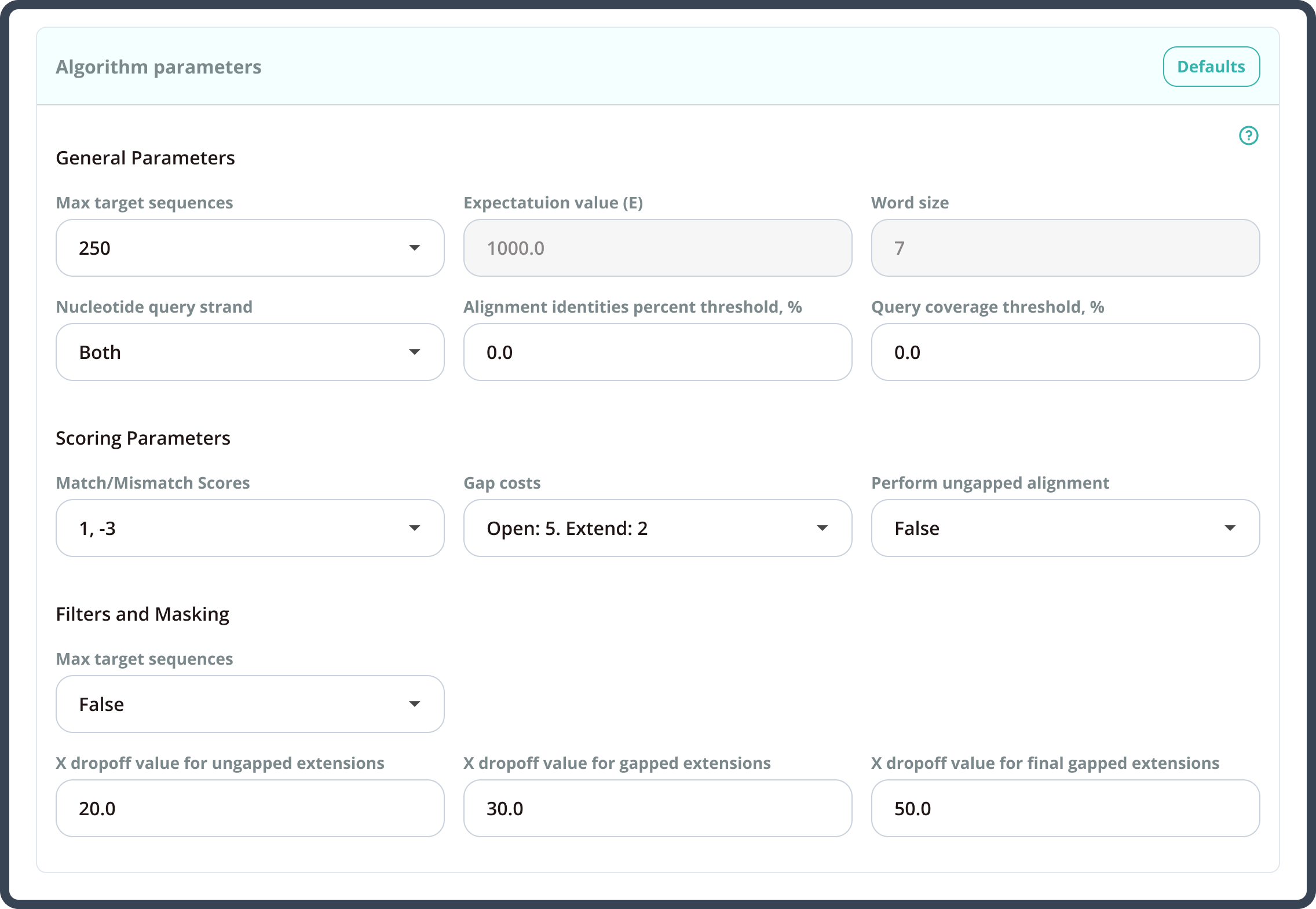
The problem lies in the fact that the matches, significant to the query sequence, are often not found under the standard BLAST settings. Being a probability based algorithm, BLAST omits a considerable amount of sequences when conducting a sequence search with a short query sequence (less than 20 bases). The reason for this is that the significance threshold is set too stringently, and the default sequence length parameter is set too high. Thus,BLAST was failing to provide the necessary level of accuracy to the researchers.
The client was in need of BLAST algorithm customizations to gather more accurate results from searches with short query sequences.
By having strong technological skills and deep domain knowledge in bioinformatics, our development team elaborated the algorithm parameters and additional search filters, which specifically address the issues of the searches with short query sequences. The system was redesigned to automatically adjust these developments: the preset values on the search launch page help to provide optimum results.
The Azati engineering team implemented the changes to the extensive BLAST algorithm code,thus making the system highly responsive to the client’s research needs.
SOME DETAILED INFORMATION NOT DISCLOSED DUE TO NDA RESTRICTIONS
If you are interested in the development of a custom solution — send us the message and we'll schedule a talk about it.
JavaScript, Ruby
HR Planning SoftwareThe customer asked Azati to audit the existing solution in terms of general performance to create a roadmap of future improvements. Our team also increased application performance and delivered several new features.
Python
Stock Market Trend Discovery with Machine LearningAt Azati Labs, our engineers developed an AI-powered prototype of a tool that can spot a stock market trend. Online trading applications may use this information to calculate the actual stock market price change.
Python
Semantic Search Engine for Bioinformatics CompanyAzati designed and developed a semantic search engine powered by machine learning. It extracts the actual meaning from the search query and looks for the most relevant results across huge scientific datasets.
Java, JavaScript
E-health Web Portal for International Software IntegratorAzati helped a well-known software integrator to eliminate legacy code, rebuild a complex web application, and fix the majority of mission-critical bugs.
JavaScript, Ruby
Custom Platform for Logistics and Goods TransportationAzati helped a European startup to create a custom logistics platform. It helps shippers to track goods in a real-time, as well as guarantees that the buyer will receive the product in a perfect condition.