Drop us a line
If you are interested in the development of a custom solution — send us the message and we'll schedule a talk about it.
Identifying and fixing the bottleneck for reducing the client’s program execution time. With this improvement, the client achieved a 1,000x reduction of an embedded algorithm execution time and 80x reduction for the whole client’s software.
On the basis of genetic engineering a whole branch of the pharmaceutical industry, called the “DNA industry”, has emerged and it is one of the modern branches of biotechnology. More than a quarter of all medicines currently used in the world contain ingredients from plants. Genetically modified plants are a cheap and safe source for obtaining fully functional medicinal proteins (antibodies, vaccines, enzymes, etc.) for both humans and animals.
A biotechnology customer company offers synthetic antibody products and services such as research reagents, diagnostic and biomarker discovery tools for use in drug discovery and targeted delivery for therapeutics, and bioindustrial applications.
Bioinformatics specialists are constantly dealing with huge amounts of data (i.g. the results of the genome sequencing just for one person occupy about 100 gigabytes). Therefore, the processing of such massive data requires Data Science approaches and tools.
That is why the customer turned to the Azati team with the main task of performance enhancement.
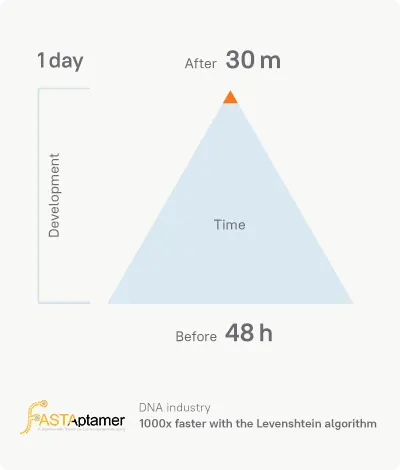
In the process of DNA sequencing, the client receives lots of data. Previously, it took 48 hours for the client’s software to process the data. The company turned to Azati with a request for increasing their software performance within a short period.
Client’s software processes data in multiple steps; on one of them it uses an outside package FASTAptamer. FASTAptamer is a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections.
It was revealed that one of the FASTAptamer’s programs — clusterization — took most of the time of the whole software performance. The great deal of time of the clusterization program, in turn, was devoted to calculating edit time (the level of similarity between the biological sequences). This calculation is based on the Levenshtein algorithm.
Although the algorithm itself is appropriate, its implementation in Perl was rather poor and took a lot of time to process the submitted sequence data. Our team decided to rewrite the code into C++ language, as the difference in programming paradigms and execution model affected the time of execution.
Our team optimized the software within 1 day. Our client ran the updated pipeline on they data that they had recently analyzed and compared the results:
Outputs were the same
1’000 quicker performance of the Levenshtein algorithm
30.5 minutes with our improvements compared to 2460.5 minutes (~48 hours) under the original software for results generation
We proposed an accelerated version of the Levenshtein algorithm and the FASTAptamer contributors added it to the official package in the subsequent release v1.0.12.
If you are interested in the development of a custom solution — send us the message and we'll schedule a talk about it.
JavaScript, Ruby
HR Planning SoftwareThe customer asked Azati to audit the existing solution in terms of general performance to create a roadmap of future improvements. Our team also increased application performance and delivered several new features.
Python
Stock Market Trend Discovery with Machine LearningAt Azati Labs, our engineers developed an AI-powered prototype of a tool that can spot a stock market trend. Online trading applications may use this information to calculate the actual stock market price change.
Python
Semantic Search Engine for Bioinformatics CompanyAzati designed and developed a semantic search engine powered by machine learning. It extracts the actual meaning from the search query and looks for the most relevant results across huge scientific datasets.
Java, JavaScript
E-health Web Portal for International Software IntegratorAzati helped a well-known software integrator to eliminate legacy code, rebuild a complex web application, and fix the majority of mission-critical bugs.
JavaScript, Ruby
Custom Platform for Logistics and Goods TransportationAzati helped a European startup to create a custom logistics platform. It helps shippers to track goods in a real-time, as well as guarantees that the buyer will receive the product in a perfect condition.